
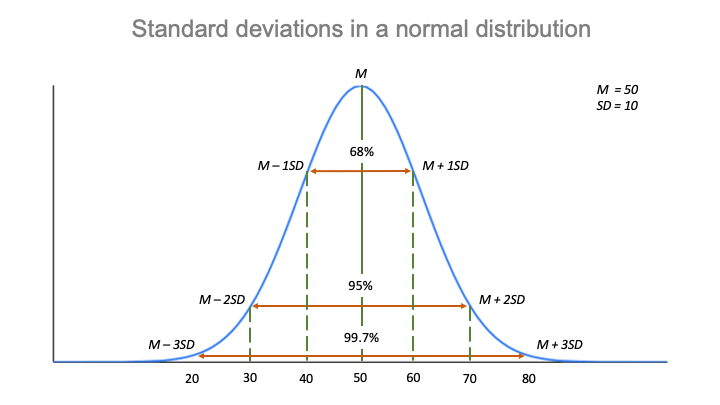
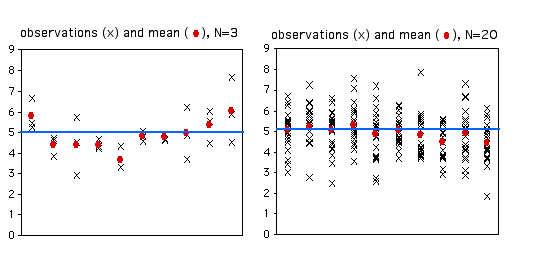

Variance-covariance matrix of these parameters, we can look at the apVar object Variance-covariance matrix of these random effects parameters. Scale, we can use the delta method and the If we wish to calculate standard errors in the standard deviation These differences can be divided by 1.96 to find the standard error in the Sd((Intercept)), and noting the symmetry of the logged interval and estimate We can see this by looking one random effect, Because standard deviations must be non-negative, the actual model-estimated value is Note that the intervals for the random effects standard deviations are NOT symmetric about theĮstimate. Reported, they can be generated using the intervals command. While the standard errors of these estimated standard deviations are not Structure: General positive-definite, Log-Cholesky parametrization Linear mixed-effects model fit by maximum likelihood Model.c <- lme(alcuse ~ coa*age_14, data=alcohol1, random= ~ age_14 | id, method="ML") Use an example dataset from Singer and Willet’s Applied Longitudinal Data Analysis.Īlcohol1 <- read.table("", header=T, sep=",") Summary command includes a section for random effects. When fitting a mixed-effects model in R using the nlme package, the information provided in the You are of your parameter values indicating how groups or subjects differ in Otherwise, these values indicate how certain The standard errors of a randomĮffects parameter, if very large, can be a red flag suggesting a problem with R presents these standard deviations,īut does not report their standard errors. Of the random intercepts or random slopes. Typically, the reported parameter of a random effect is the standard deviation Valuable information about the contribution of the random effects to the model. The standard errors of variance components in a mixed-effects model can provide


 0 kommentar(er)
0 kommentar(er)
